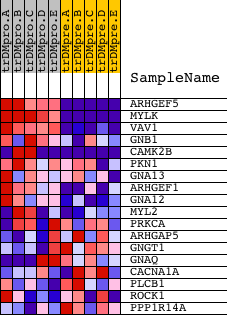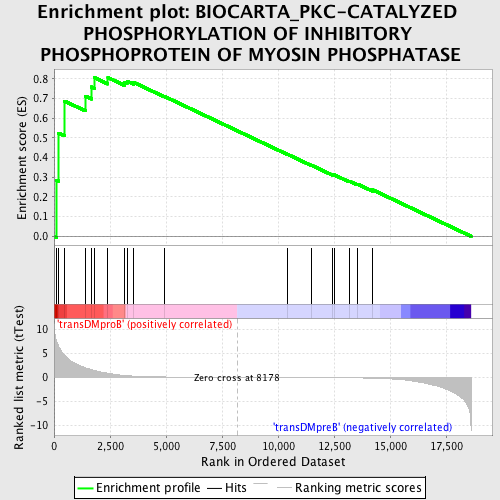
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | BIOCARTA_PKC-CATALYZED PHOSPHORYLATION OF INHIBITORY PHOSPHOPROTEIN OF MYOSIN PHOSPHATASE |
| Enrichment Score (ES) | 0.8066343 |
| Normalized Enrichment Score (NES) | 1.4826866 |
| Nominal p-value | 0.020408163 |
| FDR q-value | 0.41194376 |
| FWER p-Value | 0.999 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ARHGEF5 | 12025 | 104 | 7.577 | 0.2853 | Yes | ||
| 2 | MYLK | 22778 4213 | 200 | 6.379 | 0.5251 | Yes | ||
| 3 | VAV1 | 23173 | 461 | 4.594 | 0.6875 | Yes | ||
| 4 | GNB1 | 15967 | 1402 | 1.958 | 0.7121 | Yes | ||
| 5 | CAMK2B | 20536 | 1673 | 1.604 | 0.7592 | Yes | ||
| 6 | PKN1 | 11567 6799 | 1812 | 1.404 | 0.8057 | Yes | ||
| 7 | GNA13 | 20617 | 2402 | 0.850 | 0.8066 | Yes | ||
| 8 | ARHGEF1 | 1952 18343 3996 | 3132 | 0.392 | 0.7825 | No | ||
| 9 | GNA12 | 9023 | 3258 | 0.338 | 0.7887 | No | ||
| 10 | MYL2 | 16713 | 3560 | 0.242 | 0.7818 | No | ||
| 11 | PRKCA | 20174 | 4927 | 0.066 | 0.7109 | No | ||
| 12 | ARHGAP5 | 4412 8625 | 10426 | -0.029 | 0.4165 | No | ||
| 13 | GNGT1 | 17533 | 11492 | -0.049 | 0.3611 | No | ||
| 14 | GNAQ | 4786 23909 3685 | 12422 | -0.073 | 0.3139 | No | ||
| 15 | CACNA1A | 4461 | 12509 | -0.077 | 0.3122 | No | ||
| 16 | PLCB1 | 14832 2821 | 13167 | -0.103 | 0.2809 | No | ||
| 17 | ROCK1 | 5386 | 13538 | -0.125 | 0.2658 | No | ||
| 18 | PPP1R14A | 18310 3822 | 14216 | -0.187 | 0.2366 | No |